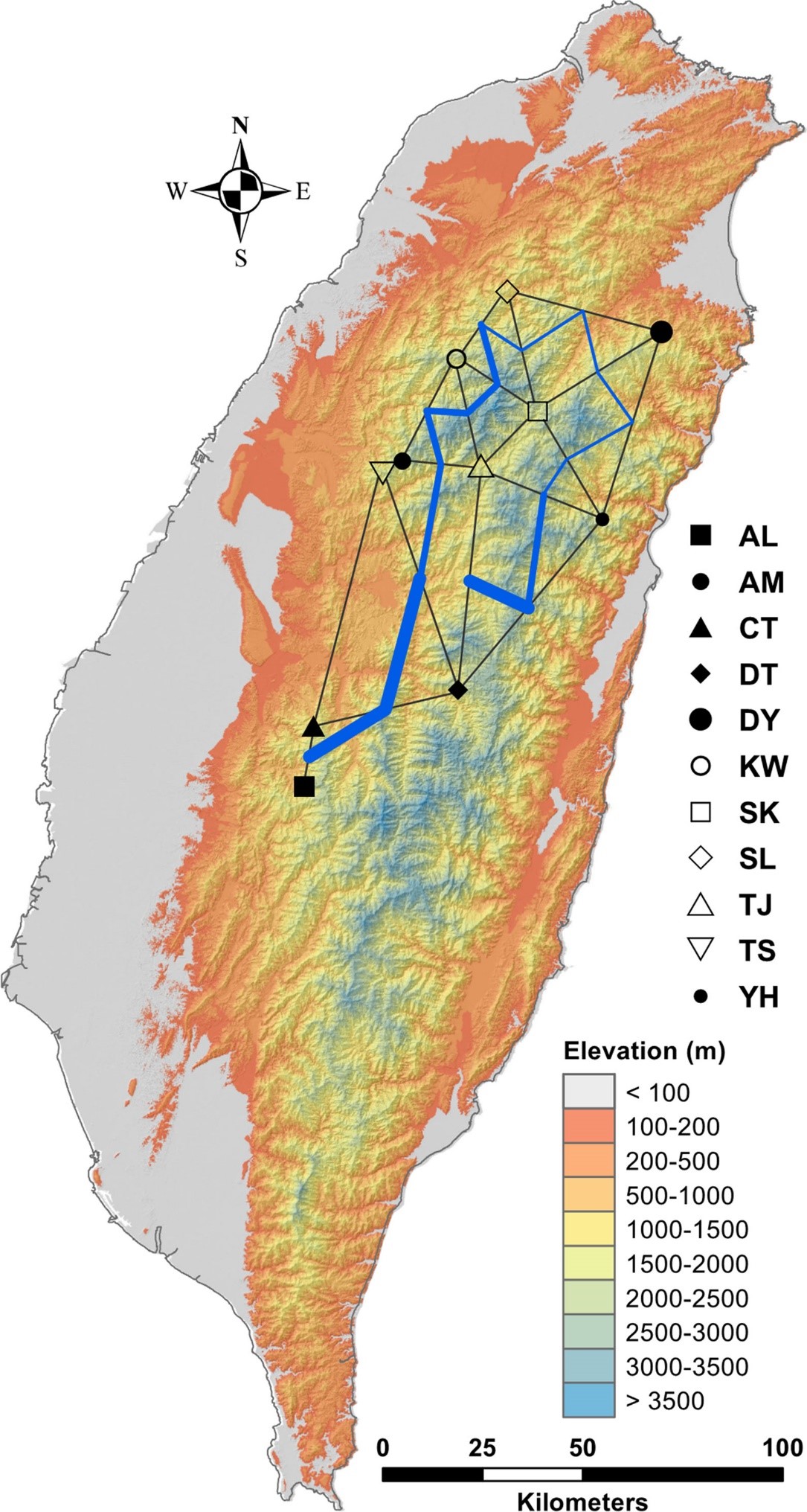
Populations can be genetically isolated by differences in their ecology or environment that hamper efficient migration, or they may be isolated solely by geographic distance. Moreover, mountain ranges across a species’ distribution area might act as barriers to gene flow. Genetic variation was quantified using amplified fragment length polymorphism (AFLP) and 13 selective amplification primer combinations were used to generate a total of 482 fragments. Here, we tested the barrier effects of mountains on the gene flow and environmentally dependent local adaptation of Cunninghamia konishii in Taiwan. A pattern of genetic isolation by distance was not found, and variation partitioning revealed that the environment explained a relatively larger proportion of genetic variation than geography. The effect of mountains as barriers to genetic exchange, despite low population differentiation indicating a high rate of gene flow, was found within the distribution range of C. konishii. Twelve AFLP loci were identified as potential selective outliers using genome-scan methods (BAYESCAN and DFDIST) and were strongly associated with environmental variables using regression approaches (LFMM, Samβada, and rstanarm) demonstrating adaptive divergence underlying local adaptation. Annual mean temperature, annual precipitation, and slope could be the most important environmental factors causally associated with adaptive genetic variation in C. konishii. The study revealed the existence of physical barriers to current gene flow and environmentally dependent adaptive divergence, and a significant proportion of the rate of gene flow may represent a reflection of demographic history.
Taiwan Island emerged during a late stage in geological history, having been formed about 5 million years ago by the collision and compression of the Philippine and Eurasian plates. It was only 3 million years ago that the present structure of Taiwan Island was formed. Such a special continental island has a high proportion of endemic species, and after the formation of Taiwan Island, most of the time was spent in the cycle of ice ages and interglacial periods. This geological history combined with climate change since the last ice age has not only influenced the history of species formation and community change, but has also resulted in a high proportion of endemism due to large habitat differences. Our research laboratory has long used Taiwan's endemic woody plant species as materials, and utilized contemporary methods of community genetics, kinship geography, and functional genetic evolution, combined with molecular evolution and bioinformatics analyses, to investigate the evolutionary history of wild woody plant species on a geologically new continental island near the mainland. The specific academic contribution of our study is that we integrate the evolution of wild woody plant species on this special island into international research trends, and provide a perspective on the evolution of plant species and community adaptation in the context of forest biodiversity in Taiwan's special region, with the hope that the international academic community can further understand Taiwan's rich endemism, unique geographical environment, and the history of the evolution of its woody plant species. It is expected that the research results of this laboratory will provide the international academic community with a better understanding of Taiwan's rich endemic species, unique geographical environment, and the evolutionary history of its woody plant species. It is expected that the results of the research laboratory will be beneficial to the development of conservation measures for Taiwan's endemic species.
In this article, we take Taiwan fir as an example to briefly introduce and analyze the possible evolution. There are three main clades of Taiwan fir: the Taiwan clade, the Yunnan-Burmese clade, and the Vietnamese clade. We explored the relative influence of neutral evolution and natural selection on the genetic divergence among these three clades. At the same time, we studied the evolution patterns of the genetic divergence among the ethnic groups of Taiwan fir growing in Taiwan using a spread by the strip length polymorphism (amplified fragment length polymorphism, AFLP) and the methylation sensitive expansion increased formability (methylation-sensitive amplification polymorphism, MSAP). These two kinds of research tools were used to analyze three Taiwan fir clades, providing a total of 102 samples for genetic polymorphism analysis. For the 1,413 AFLP and 462 MSAP bands, we analyzed the genome scan, broad linear patterns, and broad linear admixture patterns, and found that the genetic motifs that may be subject to natural selection were detected only in the comparison of the three clades of F. Taiwanensis, and their proportions were low. In other words, most of the genetic and epigenetic variation is neutral, and may be the result of opportunistic drift. No genetic motifs subject to natural selection were detected among the local Taiwan fir groups, indicating that the divergence among these Taiwan fir groups is mainly a neutral evolutionary pattern. The genomes of the three Taiwan fir clades that may have been subject to natural selection were analyzed by model selection and model averaging, indicating that they may be significantly associated with temperature and vegetation changes.
In addition, the genomic single nucleotide polymorphism (SNP) markers can be obtained by ddRADseq (double digest Restriction Associated DNA sequencing), which can effectively study species differentiation, community structure, and detect natural selection markers. In this study, we investigated the use of ddRADseq to distinguish the genetic heritage of the Taiwan Oleander community and its species from its ancestral species, D. viridis. The results of the community genetic analysis were examined in terms of the percentage of missing value to determine which of the three sets of values, 40%, 50%, and 60%, could be more appropriately used for the analysis of this study. Based on the results of the analysis, we used 50% missing value for the species differentiation and community genetic analysis. The results of the sample differentiation and kinship evolution tree analysis showed that the samples of F. taiwanensis and F. taiwanensis could be clearly distinguished from each other, and the genetic relationships between the samples of F. taiwanensis and the communities could be effectively differentiated. However, 30 forest compartment ethnic groups belonging to Taiwan Keteleeria DAWU have few samples from northern obvious case of gene exchange with 23 SNPs that were detected with a significant deviation from neutral evolution of genetic differentiation values. Fifteen of them were significantly correlated with environmental factors by logistic regression analysis. This result showed that the genetic divergence among Taiwanese Abies populations was related to the differences in environmental factors, showing the divergence pattern of isolation by environment (IBE), and the multi-matrix random regression analysis showed that in addition to significant IBE, there was also a clear pattern of isolation by distance (IBD). The genetic divergence pattern among the 23 genetically divergent groups was also evident. Among the 23 SNPs with genetic divergence, three SNPs were found to be related to the maintenance of granulosa metabolic homeostasis and stress tolerance by BLASTN, which also indicated that genetic divergence among populations in Taiwan is related to adaptive evolution in response to habitat environment.











